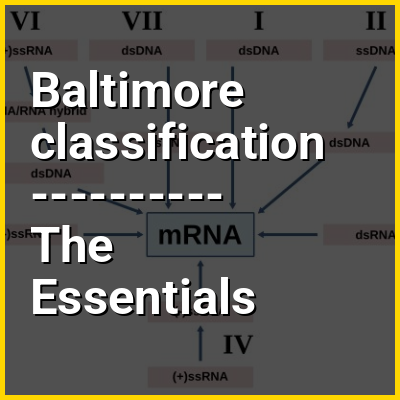
Baltimore classification is a system used to classify viruses by their routes of transferring genetic information from the genome to messenger RNA (mRNA). Seven Baltimore groups, or classes, exist and are numbered in Roman numerals from I to VII. Groups are defined by whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, whether a single-stranded RNA genome is positive-sense (+) or negative-sense (–), and whether the virus makes DNA from RNA (reverse transcription (RT)). Viruses within Baltimore groups typically have the same replication method, but other characteristics such as virion structure are not directly related to Baltimore classification.
The seven Baltimore groups are for double-stranded DNA (dsDNA) viruses, single-stranded DNA (ssDNA) viruses, double-stranded RNA (dsRNA) viruses, positive-sense single-stranded RNA (+ssRNA) viruses, negative-sense single-stranded RNA (–ssRNA) viruses, ssRNA viruses that have a DNA intermediate in their life cycle (ssRNA-RT), and dsDNA viruses that have an RNA intermediate in their life cycle (dsDNA-RT). Only one class exists for ssDNA viruses because their genomes are converted to dsDNA before transcription regardless of sense. Some viruses belong to more than one Baltimore group, such as DNA viruses that have either dsDNA or ssDNA as their genome.